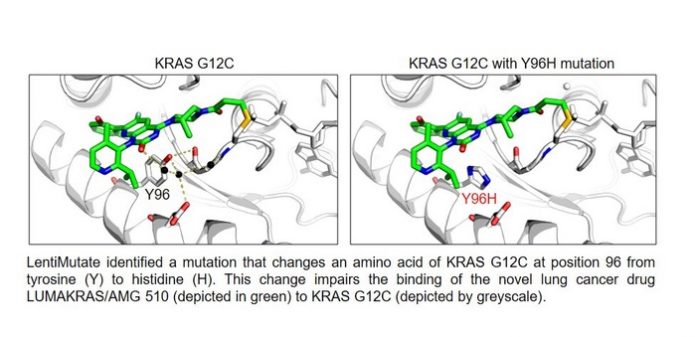
Using a virus to purposely mutate genes that produce cancer-driving proteins could shed light on the resistance that inevitably develops to cancer drugs that target them, a new study led by UT Southwestern scientists suggests. The findings, published online in Cancer Research, could help researchers develop drugs that circumvent resistance, validate new drug targets, or better understand the interaction between drugs and their target proteins.
“We believe this approach will be a very useful tool in the fight against cancer therapeutic resistance and could have potential in a variety of other areas of drug development,” said study leader Ralf Kittler, Ph.D., Associate Professor of Pharmacology in the Eugene McDermott Center for Human Growth and Development and the Harold C. Simmons Comprehensive Cancer Center. Dr. Kittler co-led the study with John D. Minna, M.D., Professor of Internal Medicine and Pharmacology, Director of the Hamon Center for Therapeutic Oncology Research and member of the Simmons Cancer Center.
Targeted therapies represent a major advance in cancer treatment for multiple tumor types, comprising drugs that specifically alter the function of oncoproteins that drive tumors to grow and spread. They are often oral agents with low toxicity that provide symptom relief and prolong survival. However, explained Dr. Kittler, these drugs have a marked drawback: they lose effectiveness over time as tumors become resistant because the genes responsible for the targeted oncoproteins inevitably mutate, producing proteins that no longer bind the drugs. For example, patients with non-small cell lung cancer are often treated with drugs that inhibit a protein known as the epidermal growth factor receptor (EGFR) providing great clinical benefit; unfortunately, most of these tumors develop resistance to the treatment within about a year. This response has led to second-, third-, and even fourth-generation versions of such EGFR-targeting drugs to try to overcome this resistance.
Although methods exist to predict mutations that will develop in cancer target genes – an important step toward developing drugs that can attack the resulting mutant proteins – these methods are cumbersome, expensive, time-consuming, or can only predict a limited type of mutation known as a point mutation, Dr. Kittler explained.
Looking for a better way to predict therapeutic resistance, the researchers developed a technique they call LentiMutate. This approach relies on a class of viruses called lentiviruses to cause mutations. In contrast with human cells and many other viruses, lentiviruses take RNA and convert it to DNA while infecting its target cells to eventually produce proteins; however, this process is inherently error-prone, producing mutant mistakes in the resulting DNA.
Working with a lentivirus engineered to make it even more error-prone, Dr. Kittler and his colleagues used the vector to insert EGFR RNA in human cells, causing the cells to produce mutant versions of this protein. They then dosed the cells with a commonly used inhibitor for EGFR called gefitinib to search for resistant cells. By sequencing the introduced single gene in the resistant cells, the researchers were able to identify several mutations that made EGFR resistant to gefitinib, a first-generation anti-EGFR drug, including those previously identified in human patients.
Further experiments showed that LentiMutate was able to identify mutations that conferred resistance to the fourth-generation anti-EGFR drug osimertinib, which is now the standard of care for EGFR mutant non-small cell lung cancer. The approach also identified mutations that cause resistance to imatinib, a drug that targets the BCR-ABL1 protein, which drives chronic myelogenous leukemia, and AMG 510, a drug that targets a specific mutant form of the KRAS protein, which drives non-small cell lung cancer.
Dr. Kittler noted that identifying these mutations through LentiMutate can greatly speed up the process of developing new drugs that can bind to the drug-resistant mutant proteins so that it takes weeks rather than years. LentiMutate could also be used in different ways in drug development: to confirm that new drugs are acting on the target protein and not a different one, to help researchers gain a better understanding of how drugs are interacting with their targets, or to develop new types of drugs for a variety of other diseases beyond cancer.
“Precision medicine that comes from sequencing a patient’s tumor to identify specific proteins to target for therapy has revolutionized cancer treatment. However, we need patients to be cured and not just benefit for 10 to 15 months from such targeted therapy,” said Dr. Minna. “To do this, we need to deal with drug resistance mutations, including by developing new drugs, and LentiMutate gives us an important new tool in our research armamentarium to help solve this pressing problem.”
Other UTSW researchers who contributed to this study include Paul Yenerall, Rahul K. Kollipara, Kimberley Avila, Michael Peyton, Yan Liu, and Kenneth D. Westover.
A patent pending for LentiMutate lists Yenerall, Dr. Minna, and Dr. Kittler as inventors. Dr. Minna receives licensing royalties from the National Cancer Institute and UT Southwestern for cell lines.